JUMPshiny
JUMPshiny: A User-friendly Platform for Comprehensive Analysis and Visualization of Quantitative Proteomics Data
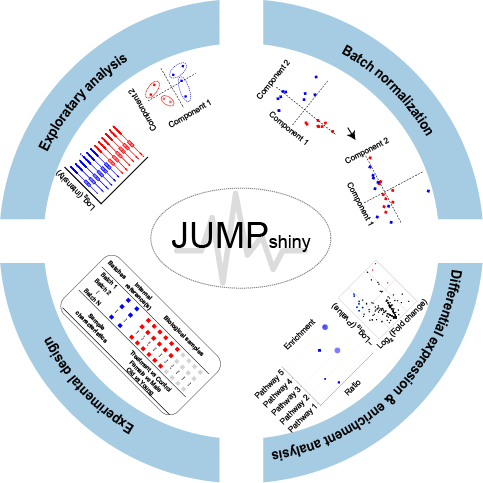
JUMPshiny is a novel, interactive, and comprehensive web-service, that is built on R-Shiny and designed for processing and presenting quantitative proteomics data. JUMPshiny includes a wide range of visualizations and offers a streamlined workflow, including experimental design, data exploration, batch normalization, differential analysis, and enrichment analysis.
SMAP
SMAP is a pipeline for sample matching in proteogenomics
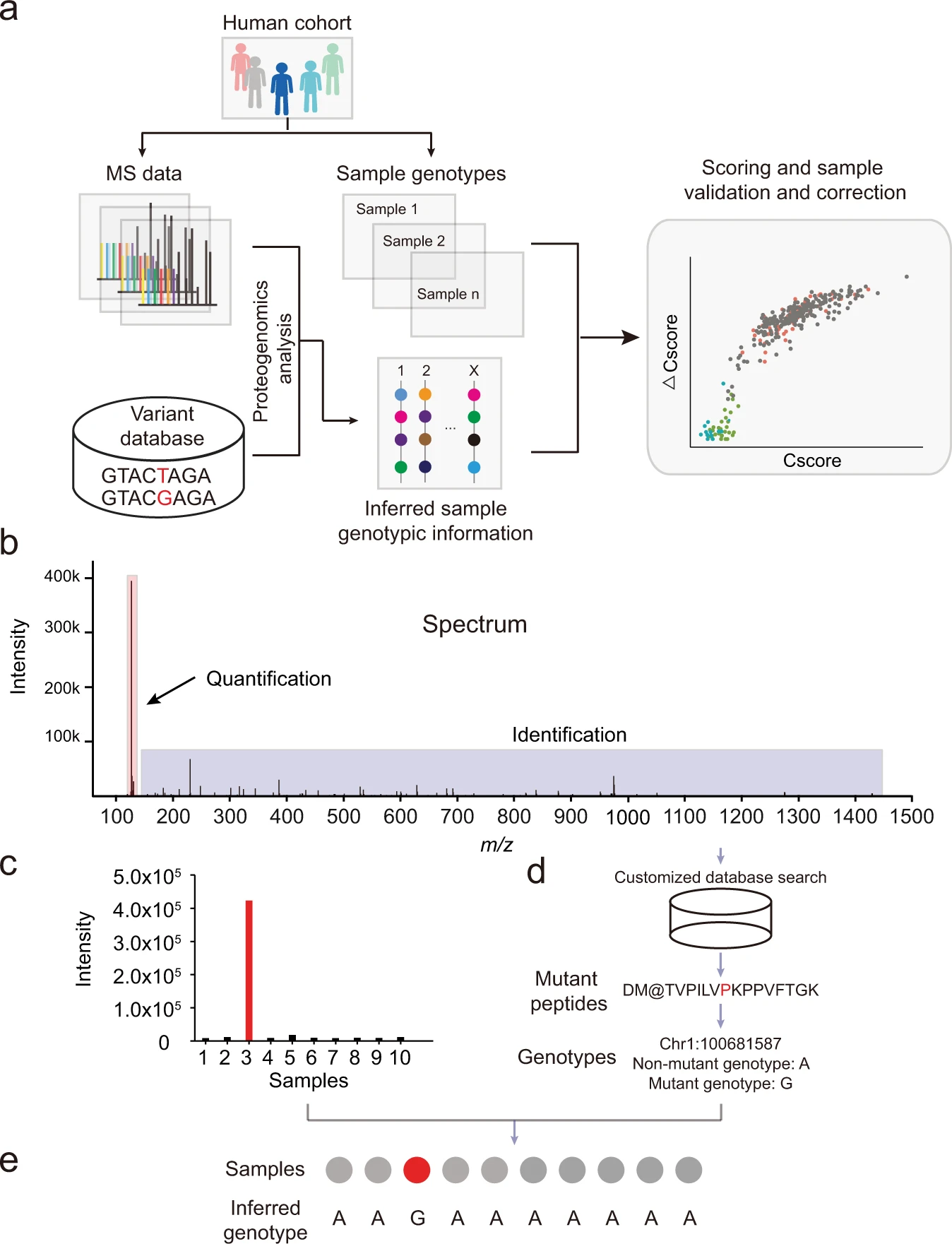
SMAP is a pipeline to validate and correct sample identity based on a combination of concordance and specificity scores. SMAP first detects variant peptides from multiplexed isobaric labeling-based quantitative proteomics data using the proteogenomics approach. Then it infers allelic information for each sample based on its expression level of the variant peptides. This shiny app allows users to input, visualize and filter their data interactively, run SMAP on their filtered data, and export the results! You can visit the GitHub or the Google webpage for additional information on running SMAP from the command line.
JUMPsem
JUMPsem: A computational tool to infer enzyme activity using post-translational modification profiling data
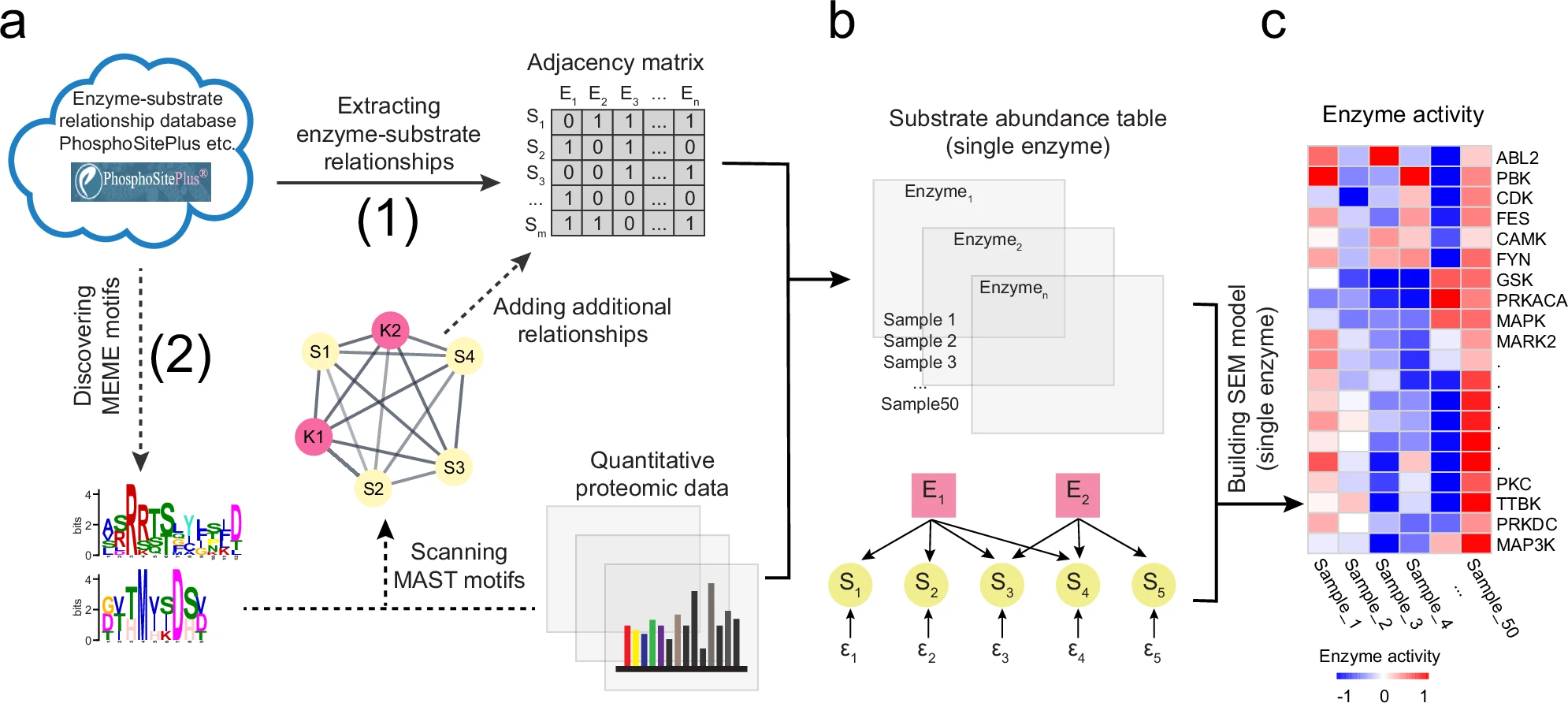
JUMPsem is a scalable and publicly available tool for inferring protein enzyme activities, potentially facilitating targeted drug development. Users could use the package from GitHub or access the online R shiny webplatform through the button below.
Wang Lab@2025
Proudly powered by WordPress and designed by Aijun Zhang
